Pictured above are some Kalash children. You notice in the foreground and center a child who could easily pass as European and draw no notice on the streets of Gdansk, Poland. But look at the child right behind her, I would guess she’d draw no notice on the streets of New Delhi!
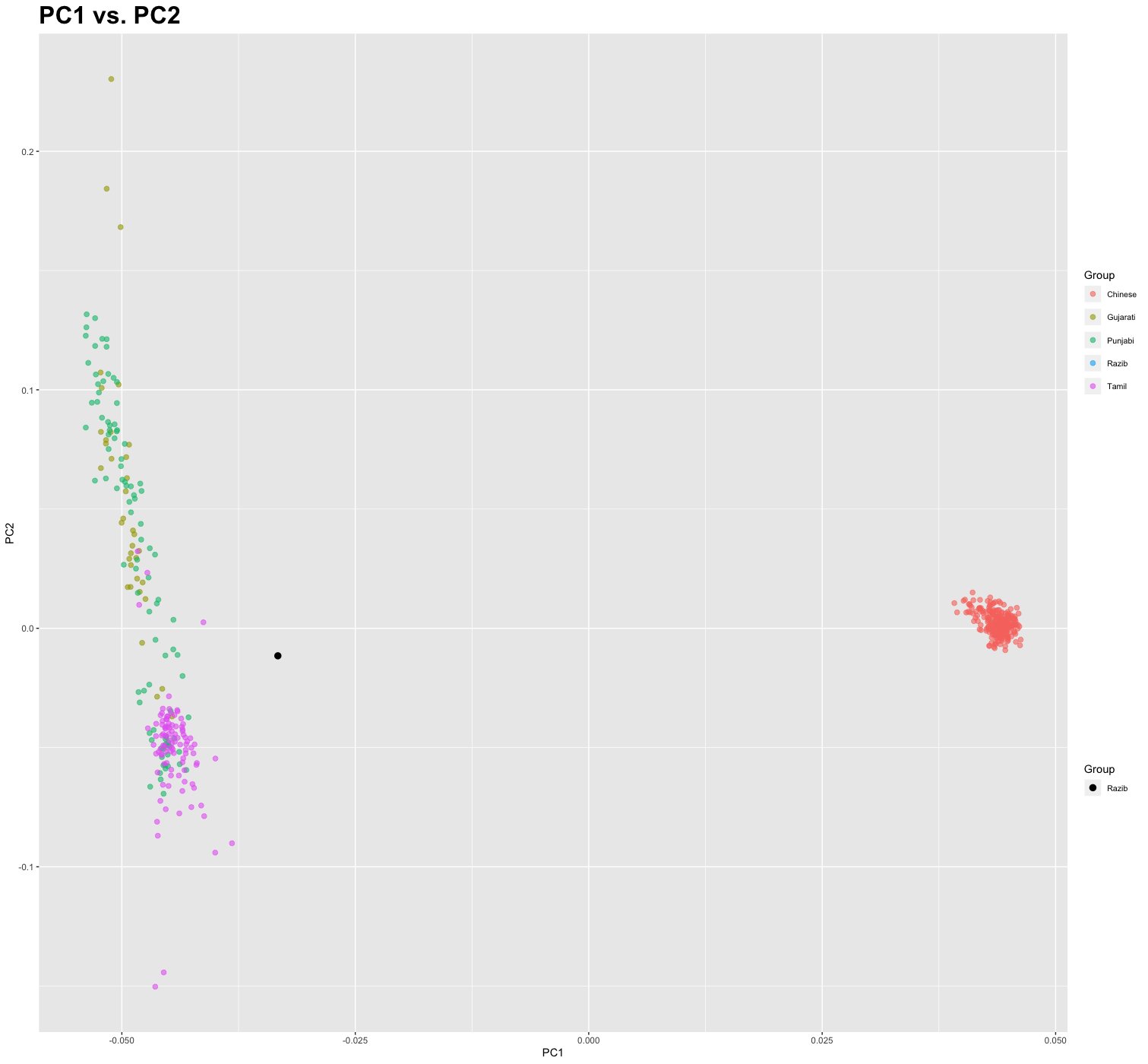
Though the Kalash are noted for their fair features, most of them look more West Asian than anything else, and from what I can tell as many have a “northwest Indian” phenotype as a “European” one. Genetically we know that they are good proxies for “Ancestral North Indians” (ANI). About ~30% of their ancestry can be modeled as derive from the steppe peoples, such as the Sintashta. Indo-Aryans. The other ~70% of their ancestry is similar to that of the Indus Valley Civilization (IVC) people, which itself can be decomposed as mostly ancient Southwest Eurasian-adjacent (i.e., derived after the Last Glacial Maximum from the ancestors of Zagros farmers) and a minority of ancestry that is more like that of Andaman Island and pre-Neolithic Southeast Asians (“Ancient Ancestral South Indians,” or AASI).
 Another thing to note about the Kalash is that they are genetically very homogeneous. This is due to the fact that they live in an isolated region, and their non-Muslim religion means that they have not intermarried with nearby Muslim people. What does this imply? It means that the Indian-looking girl is exactly the same ancestrally as the European-looking girl. Both have the same proportion of AASI and Indo-Aryan ancestry. That being said, the Indian-looking girl exhibits features more like that the AASI than the European-looking girl. Why?
Another thing to note about the Kalash is that they are genetically very homogeneous. This is due to the fact that they live in an isolated region, and their non-Muslim religion means that they have not intermarried with nearby Muslim people. What does this imply? It means that the Indian-looking girl is exactly the same ancestrally as the European-looking girl. Both have the same proportion of AASI and Indo-Aryan ancestry. That being said, the Indian-looking girl exhibits features more like that the AASI than the European-looking girl. Why?
The simple reason is that the genes which vary and encode salient physical features are a much smaller subset than the total genome. Therefore, they are subject to much higher variance from individual to individual (lower N in the denominator).
Here’s a concrete example. Compare eye color to inferring total ancestry and your total ancestry. Modern SNP-array ancestry inference relies on 100,000 to 1 million genomic positions. It is pretty good as a proxy for the 10 to 100 million SNPs out of your 3 billion base pairs that define your variable ancestry. For eye color, there are a few dozen genes at most, and more honestly a handful that really impacts variation. For Europeans, 75% of the variation of blue vs. non-blue eye color is due to variation around one genetic region, the HERC2-OCA2 locus. This means that just because someone has blue eyes, one can’t be sure that one has much European ancestry at all!
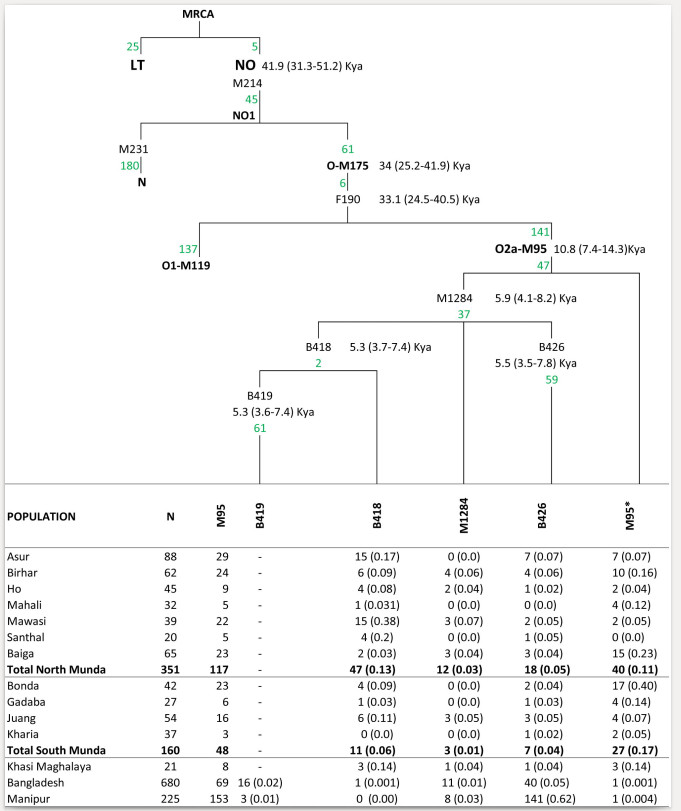
In the 1000 Genomes South Asian populations the SNPs for “blue eyes” are 2 to 10% frequency by population. Since the expression is recessive (you need both copies of the “blue eye” variant), assuming just this SNP you’d expect 0.05% to 1% manifestation of the characteristic in Indian-origin populations. The people with blue eyes have no more or less European ancestry than anyone else in their family.
Where does this leave us? You should understand from this that within a given family or ethnic group there is going to be a range of appearances, and a range is normal within many groups without exotic ancestry. Most Bengalis have 5-20% East Asian ancestry (closer to 5 in West Bengal, closer to 20 in Comilla and Chittagong). This means most of their ancestry is South Asian, and most Bengalis look just like other Indian-origin people. But a substantial minority look somewhat East Asian, to varying degrees. This is exactly what you expect when you have a minority quantum of ancestry.
Finally, many of the commenters here made a lot of assumptions about vloggers talking about their ancestry and were quite rude. I wish you wouldn’t do that. As a matter of fact, many of the inferences may actually be correct, but you don’t know for sure, and you don’t know the whole story. I’m pretty liberal on the comments of this weblog, but if you exhibit a serial pattern of rudeness I’m going to start randomly deleting your comments (if you complain about this I will immediately ban your IP).




 In my view a level 5 PISA score is the minimum requirement for a person to be considered a high school graduate who is literate in math, able to function in the modern global economy, or be qualified to attend college. The PISA report defines a level 5 PISA score or better as a fifteen year old that “can model complex situations mathematically, and can select, compare and evaluate appropriate problem-solving strategies for dealing with them.”
In my view a level 5 PISA score is the minimum requirement for a person to be considered a high school graduate who is literate in math, able to function in the modern global economy, or be qualified to attend college. The PISA report defines a level 5 PISA score or better as a fifteen year old that “can model complex situations mathematically, and can select, compare and evaluate appropriate problem-solving strategies for dealing with them.” 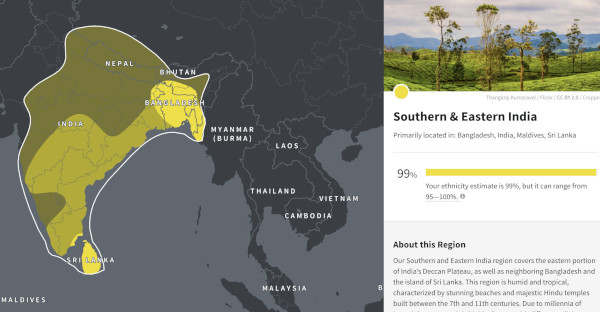
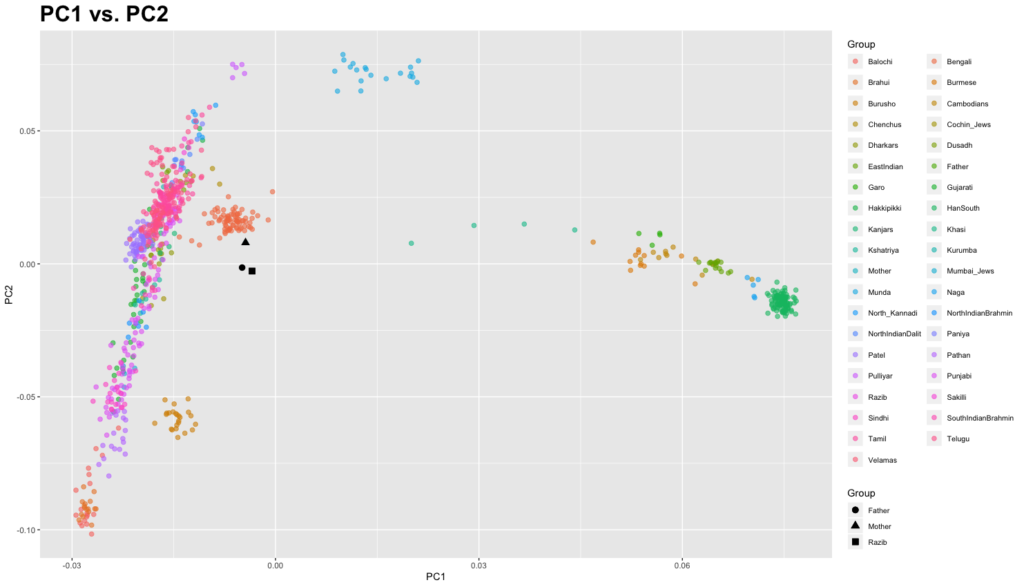
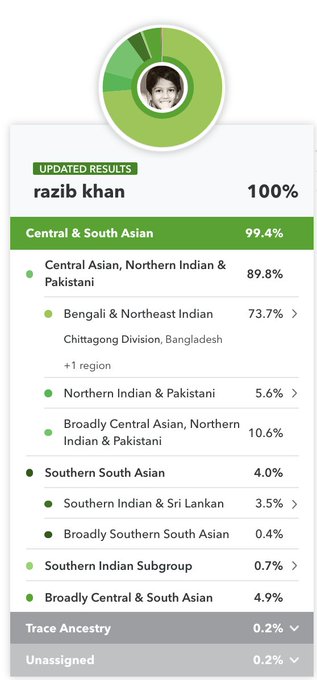 Recently I made a comment that I appreciate what 23andMe and Ancestry have done with their South Asian ancestry updates. My own results came into sharper focus. The algorithms did what they were supposed to do.
Recently I made a comment that I appreciate what 23andMe and Ancestry have done with their South Asian ancestry updates. My own results came into sharper focus. The algorithms did what they were supposed to do.