The sequences of 150,119 genomes in the UK Biobank:
We defined two other cohorts based on ancestry: African (XAF; n = 9,633; Extended Data Fig. 4) and South Asian (XSA; n = 9,252; Extended Data Fig. 5) (Fig. 3a–c). The 37,598 UKB individuals who do not belong to XBI, XAF or XSA were assigned to the cohort OTH (others). The WGS data of the XAF cohort represent one of the most comprehensive surveys of African sequence variation to date, with reported birthplaces of its members covering 31 of the 44 countries on mainland of sub-Saharan Africa (Extended Data Fig. 4). Owing to the considerable genetic diversity of African populations, and resultant differences in patterns of linkage disequilibrium, the XAF cohort may prove valuable for fine-mapping association signals due to multiple strongly correlated variants identified in XBI or other non-African populations.
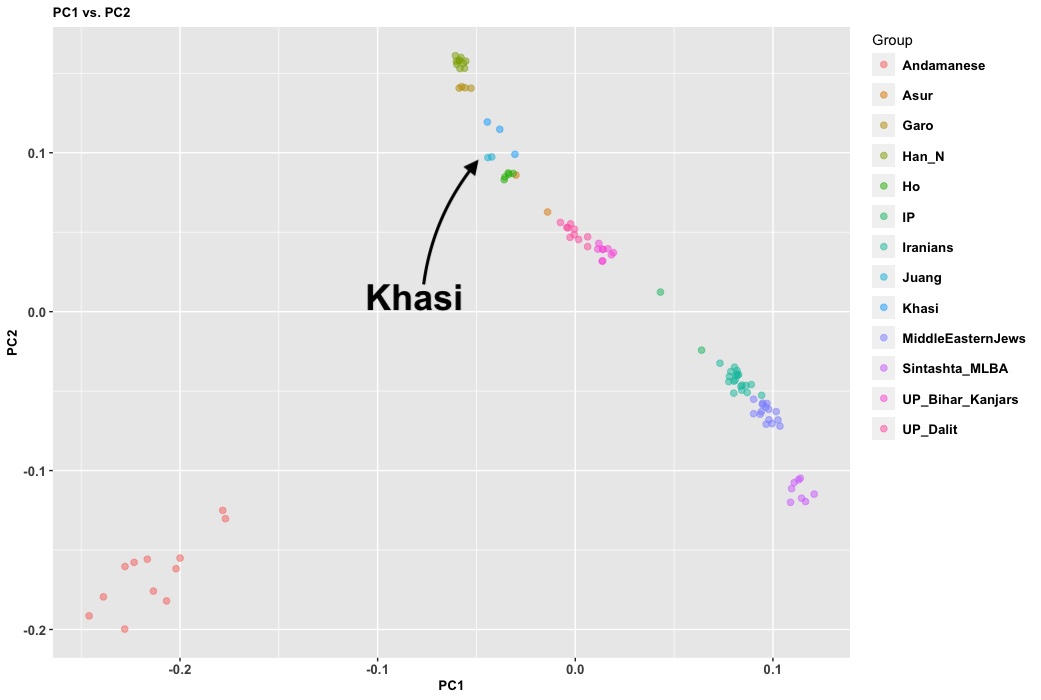
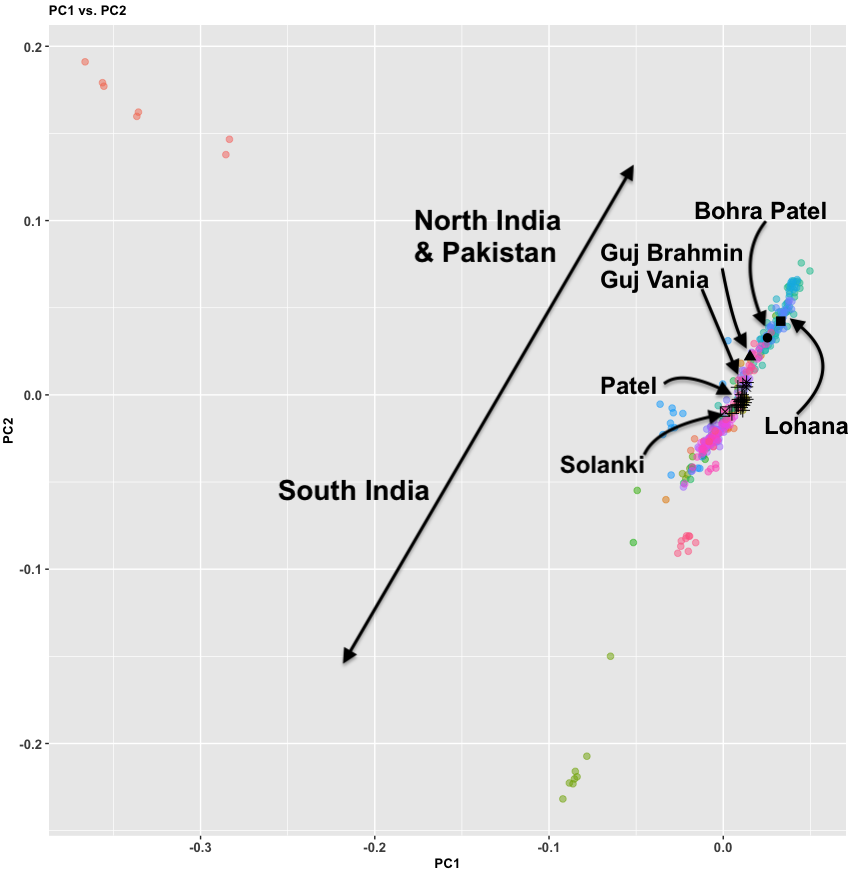
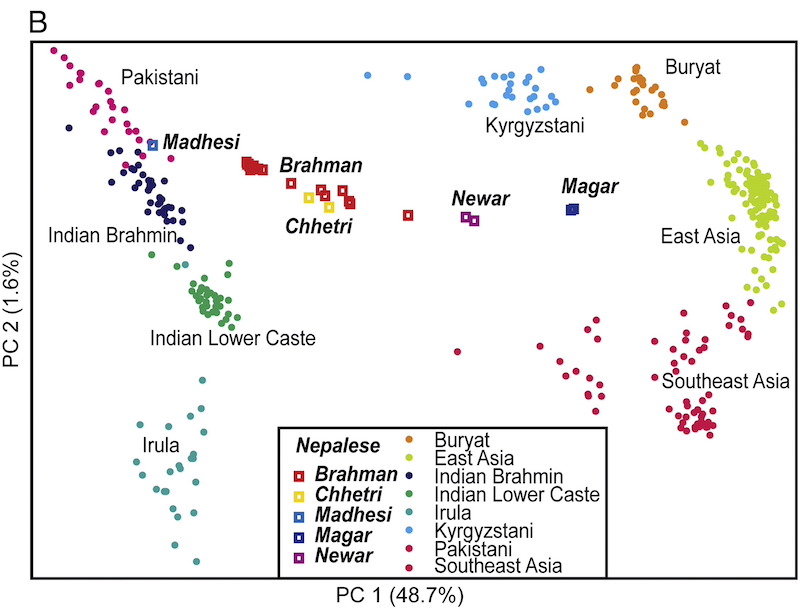
Nearly 10,000 South Asians at high-quality whole-genome sequence scale is nice to see. Obviously, this is oversampling some groups (Mirpuris, Syhletis, and East African Indians who are mostly Guju), but it’s better than nothing. It’s really sad that the British are pushing forward with this. The Chinese have started to move into sequencing their whole nation (they have millions at low coverage). This isn’t that expensive; less than $100 per person at scale. Why is India tarrying on this? I don’t have inside info but I think the Permit Raj strikes again.