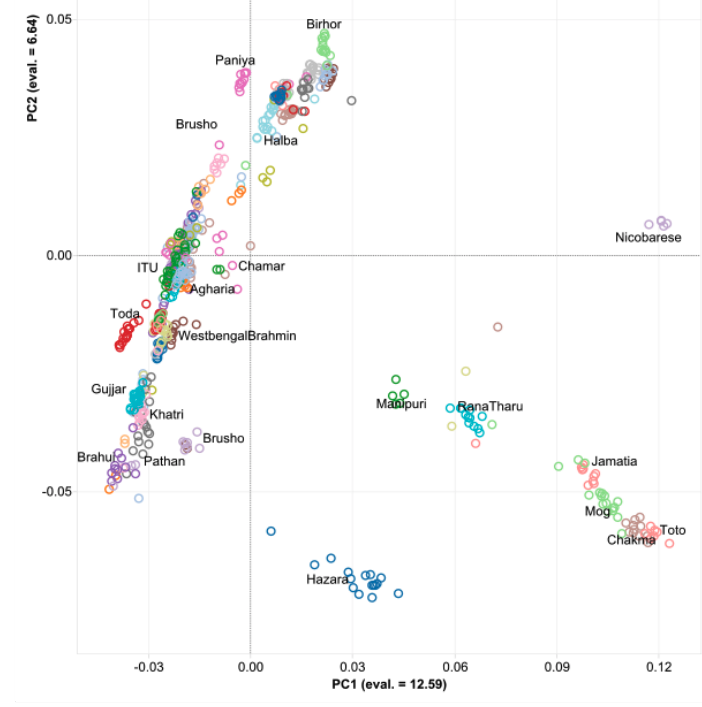
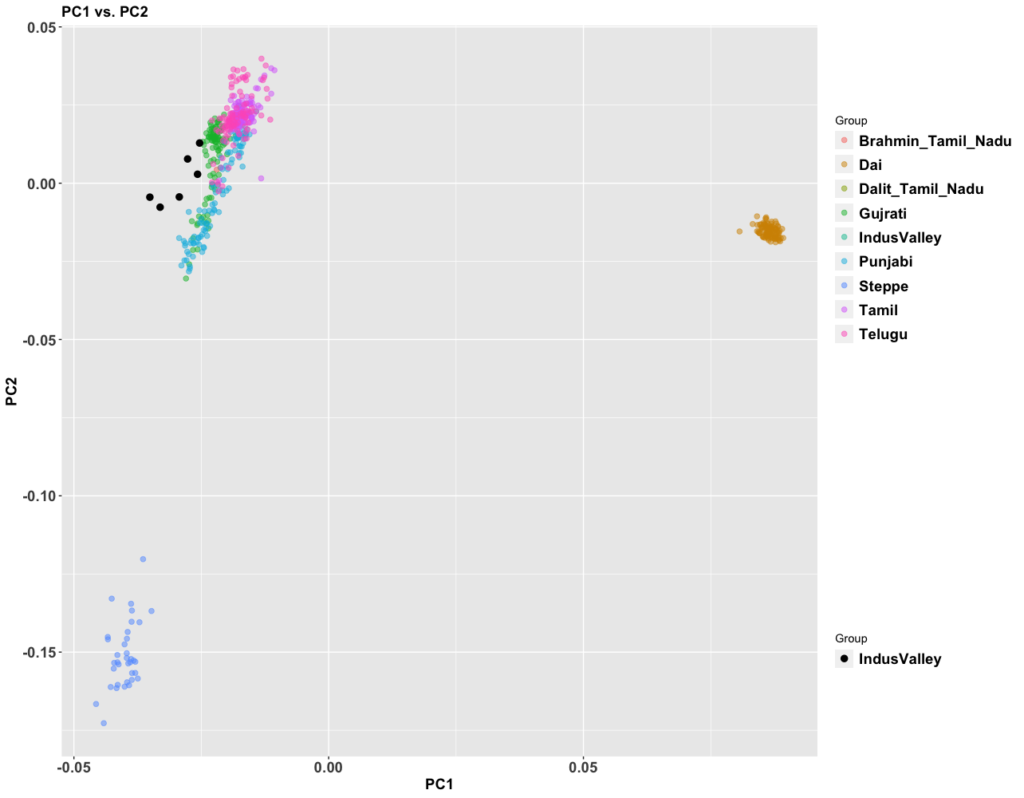
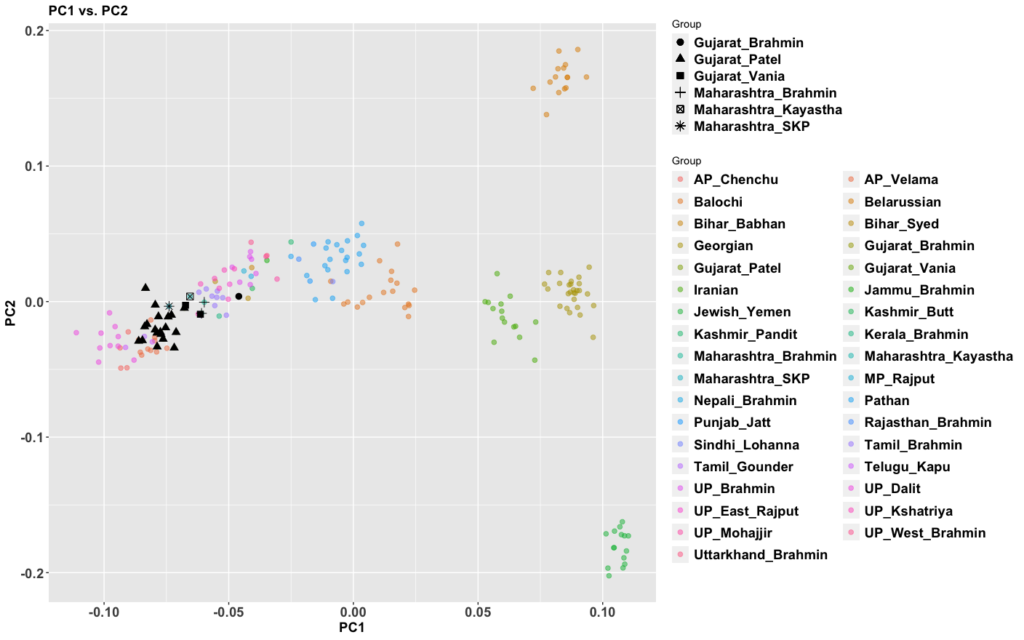
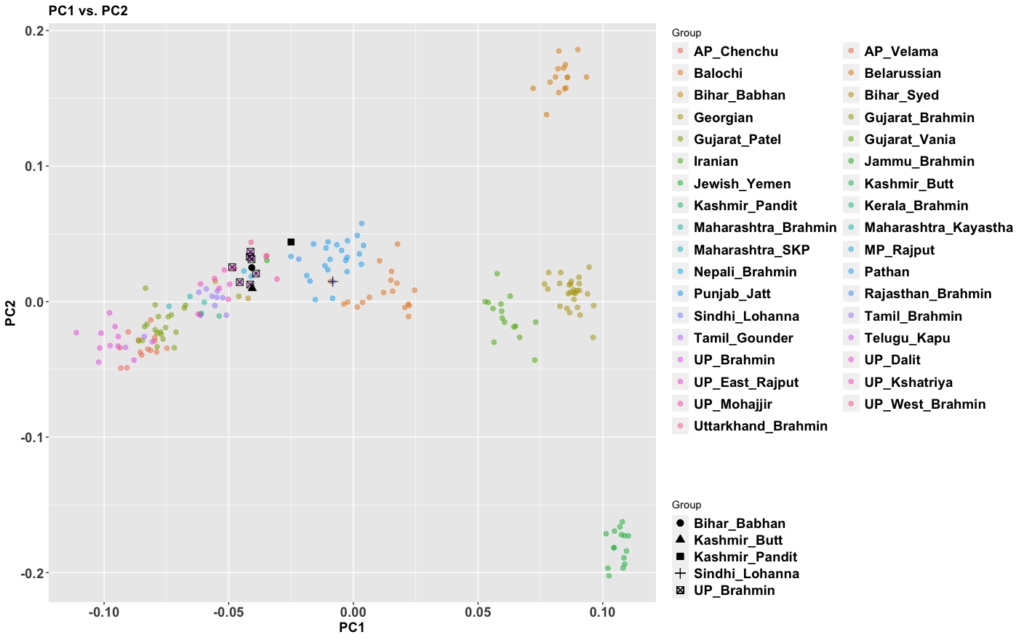
My previous post on Adivasis was not totally clear. So I’m going to try in shorter fragments and outline things so I’m more clear. I am not 100% correct with the model below (we’ll know more later), but this is my best current conception.
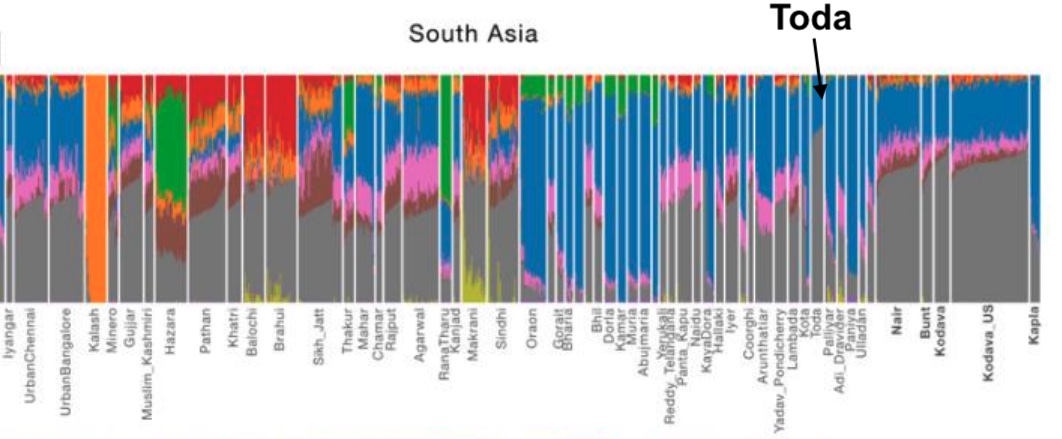
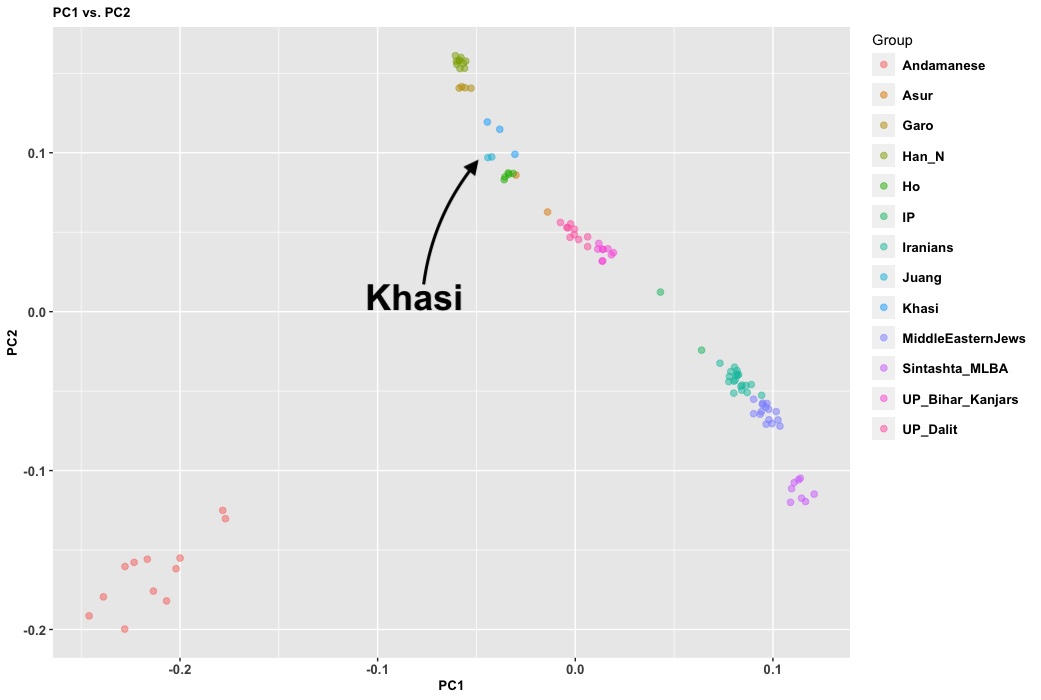
- 10,000 BC, end of the Ice Age, NW quadrant of the Indian subcontinent inhabited by a West Eurasian associated hunter-gatherers, related to the hunter-gatherers of the Zagros mountains in Iran, with some Siberian ancestry. The other three quadrants are dominated by hunter-gatherers with deep (40,000 years diverged) associations with East Eurasians and Australo-Melanesians. These “Ancient Ancestral South Indians” (AASI) seem to have separated from the Andaman Islanders (AI) more than 30-35,000 years ago, but the AI are their closest current relatives (AI-related populations were dominant in mainland Southeast Asia until 4,000 years ago, when rice farmers from southern China migrated into the region).
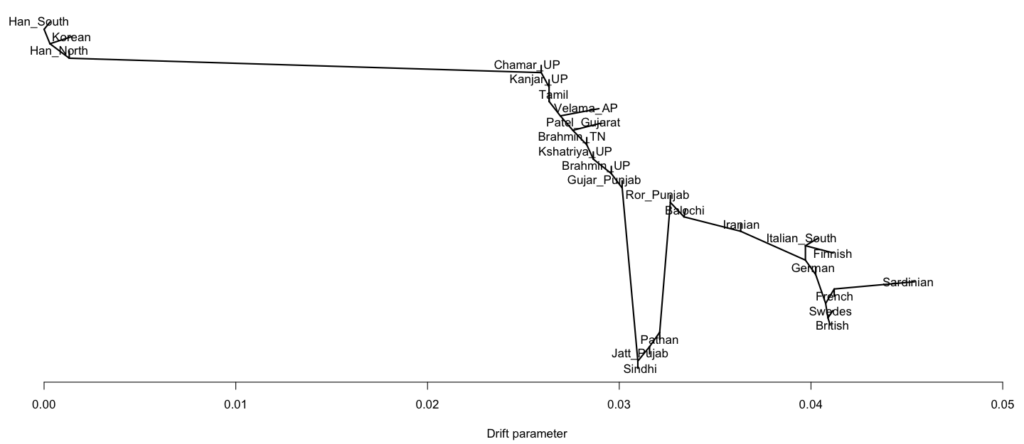
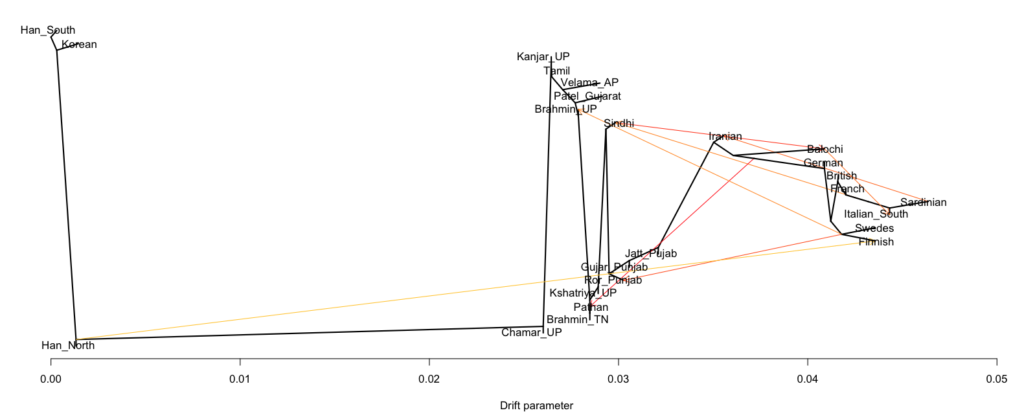
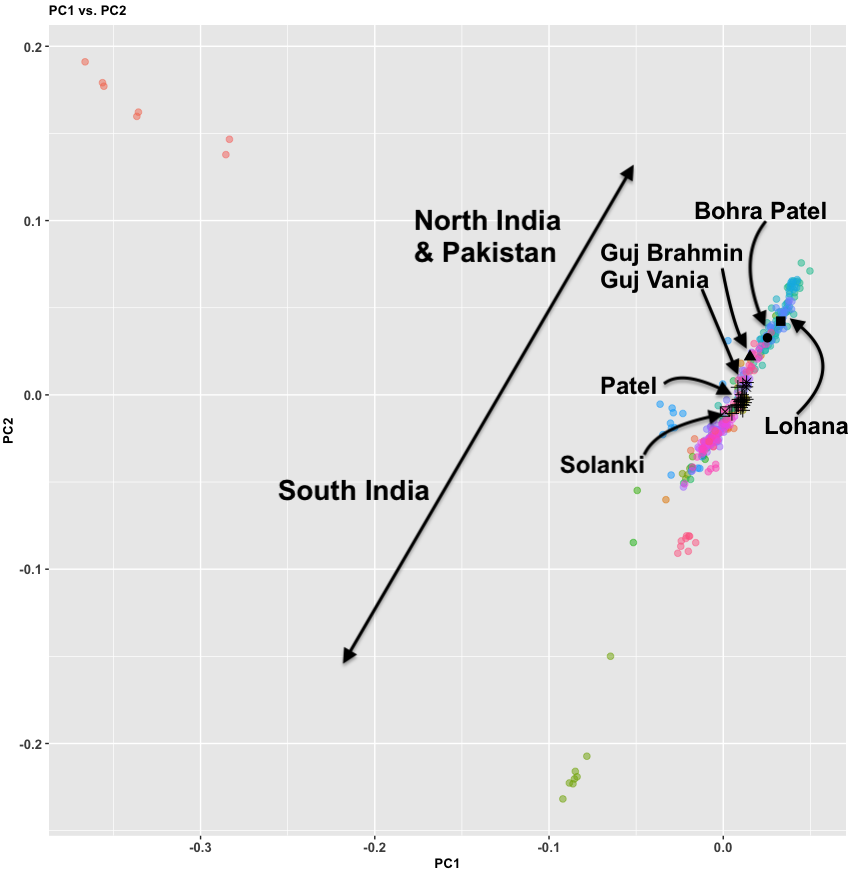
- Between 7,000 and 4,000 years ago extensive admixture occurred within the IVC zone in the NW between the IVC-Iranian-related population and AASI groups moving northwest. The resultant population was far more Iranian-related than AASI (say 10-20% AASI), and these people eventually became the “Indus Valley Civilization.
- To the south and east the AASI populations probably did experience reciprocal gene flow at the same time, as Iranian-related populations spread south and east
- Why this distinction? I believe during the late Pleistocene the Thar desert was larger and more forbidding and blocked gene flow between the easternmost West Eurasians and westernmost East Eurasians.
- Steppe ancestry likely does not show up until after 2000 BC.
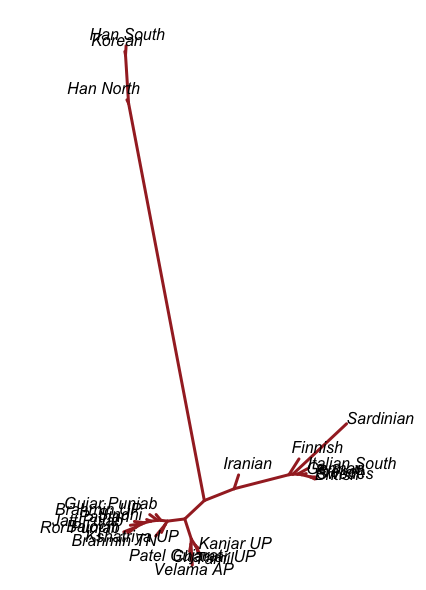
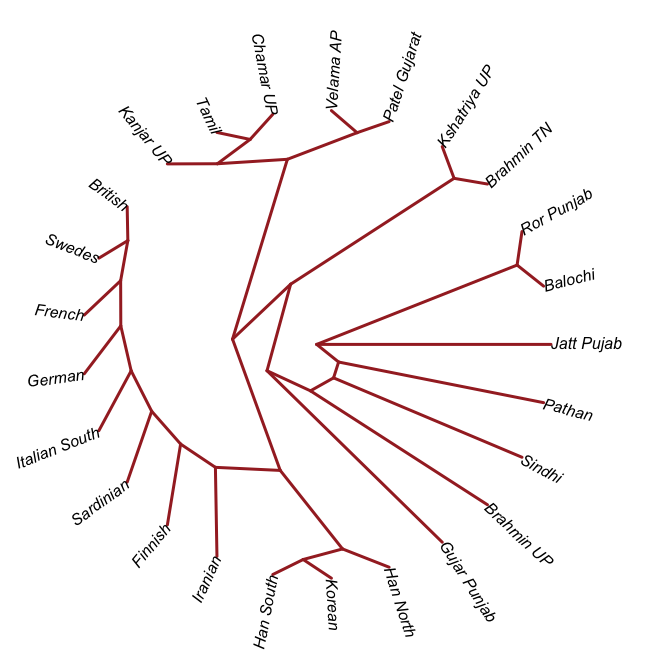
- I believe there was a Dravidian language spoken in Sindh, and later Gujarat and Maharashtra. These populations spread southward before and after 2000 BC, and eventually, they mixed with all the AASI groups in the same.
- In the period between 2000 and 1 BC there is more and more mixing and the arrival of steppe populations that become culturally ascendant across the subcontinent. In the south, the Dravidian-speaking zone, there is a distinction between post-IVC populations that engage with the expanding Indo-Aryans and those that do not engage with the Indo-Aryans
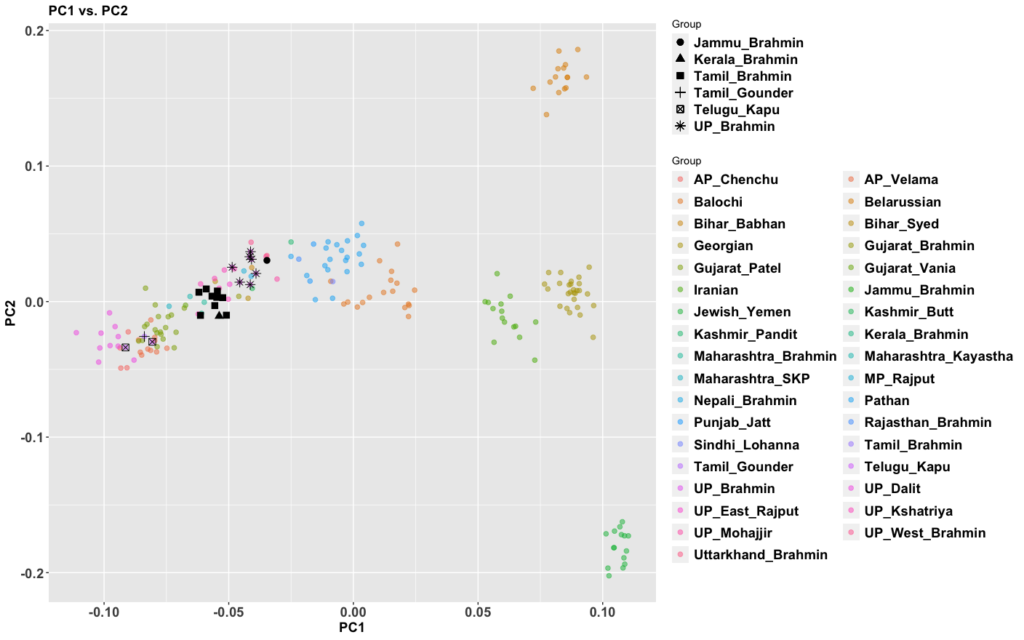
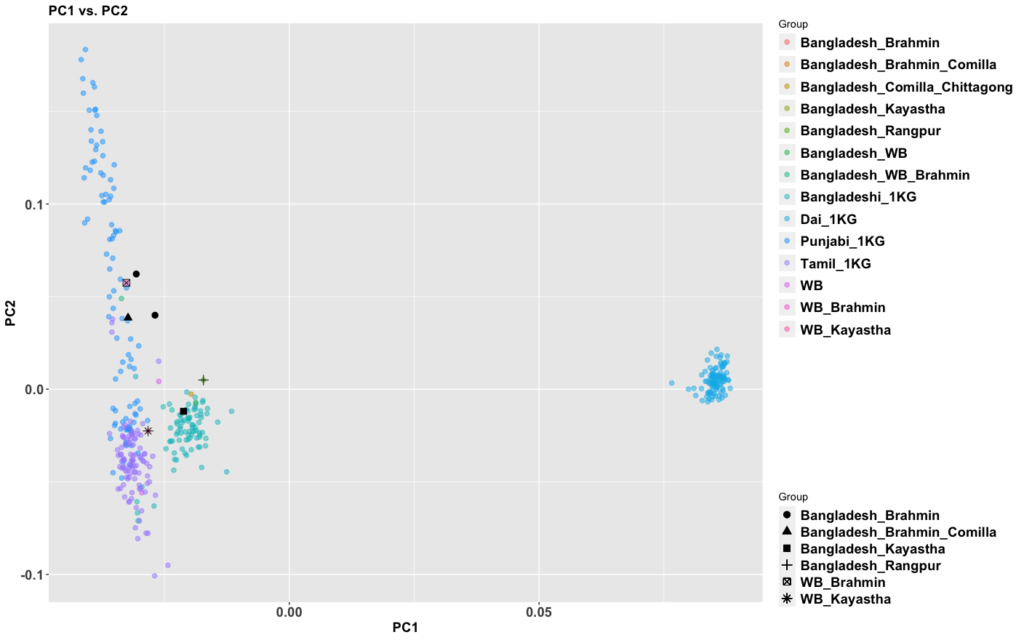
The period between 2000 and 1 BC is essential. In some areas, like the NW, large numbers of steppe people settled, and imposed their language and culture, albeit in synthesis with the local populations, who would be mostly IVC. While the IVC seems to have expanded only gingerly into the upper Gangetic plain and Gujarat, the Indo-Aryans pushed into the eastern zones, and parts of the south. The fact that Adivasi in the south have the canonically Indo-Aryan R1a-Z93 indicates that young bands of Indo-Aryan men penetrated all across the subcontinent. Their genetic imprint is clear in non-Brahmin southern groups like the Reddys, so they were ubiquitous.
But it is culture that matters more. The synthesis that developed in Punjab and Upper Gangetic plain eventually spread across the whole subcontinent and explains why Sangam literature has Sanskrit loanwords. The distinction between Adivasi and caste Hindu emerges from the distance to the expanding proto-Hindu culture based on a core of Aryan culture with indigenous accretions. This was a diverse religious and cultural matrix, but there were broad family similarities, and again, the Sangam literature alludes to “brahmins,” indicating that there was an early penetration of Aryan ritualists in the south. The Adivasi emerges not as a relict or the remnant of an early population, but as a set of societies at one of the spectra of the Aryan-indigenous synthesis that characterized the subcontinent.
The Aryan can become an Adivasi, as is attested by the Aryan men who clearly integrated themselves into those communities and lost their cultural distinctiveness. Similarly, Adivasis can become caste Hindus by adopting the norms of caste Hindus.